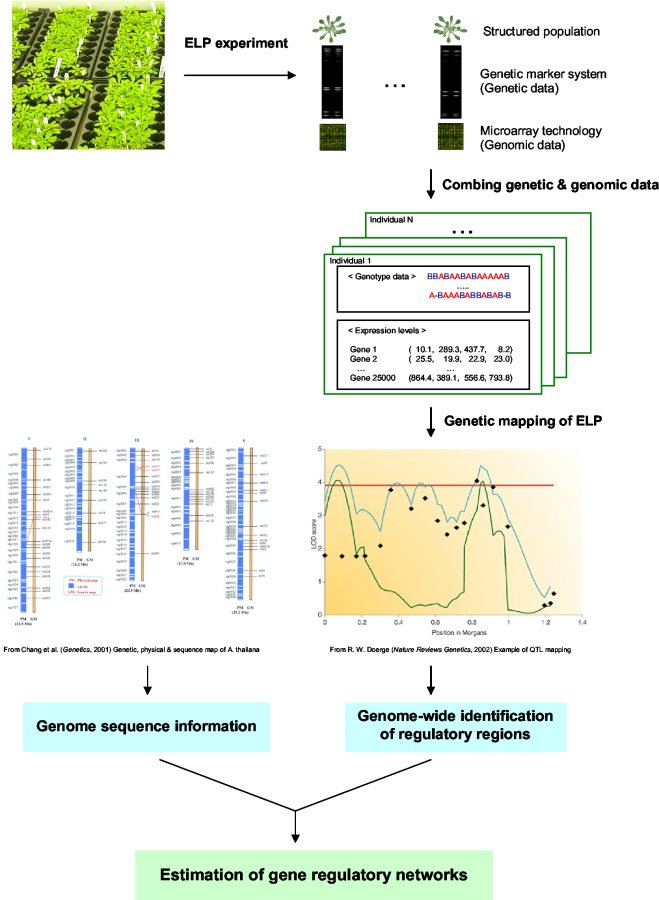
Combining QTL Analysis with Microarray Data
Expression Level Polymorphism (ELP) Analysis for Gene Regulatory Network Estimation
The molecular basis of complex traits (e.g., regulatory networks of disease resistance response) has been studied with increasing interest using various approaches, such as QTL analysis in quantitative genetics; genome sequencing, and gene expression analysis in genomics. While the underlying regulatory mechanism of complex traits has been investigated in each area of genome science, our understanding of the entire process as a whole remains a great challenge. With recent advances in biotechnology, combining information from many areas of genetics, genomics, statistics and bioinformatics, provides ways to explore the genetic and molecular architecture of complex traits at the systems biology level.
Expression level polymorphisms (ELPs) are the natural variation in induced expression patterns of genes involved in a complex trait (Doerge, 2002). Through an approach that combines an advanced QTL analysis in genetics with microarray experiments in genomics, we can identify regulatory genes/regions controlling ELPs, and examine their statistical interaction with a certain treatment that is related to regulatory pathways (Kim et al., 2005). Incorporation of the genome-wide sequence information (physical and genetic maps), available for certain organisms (e.g., Arabidopsis, mice, etc.), enables further classification of these regulatory genes into trans-acting or cis-acting regulators, and allow one to estimate putative regulatory networks of complex traits of interest.
References
Doerge, R. W. 2002. Mapping and analysis of quantitative trait loci in experimental populations. Nature Reviews Genetics, 3:43-52.
K. Kim, M. West, R.W. Michelmore, D. St. Clair, and R.W. Doerge. 2005. Old methods for new ideas: using gene expression levels to molecularly dissect complex traits. The Proceedings of the Stadler Genetics Symposium. March 2003. Edited by Perry Gustafson, Randy Shoemaker, and John Snape. Springer, NY. 89-106.
D.J. Kliebenstein, M.A.L. West, H. van Leeuwen, K. Kim, R.W. Doerge, R.W. Michelmore, D.A. St.Clair. 2006. Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics. 172: 1179-1189.
M.A.L. West, H. van Leeuwen, A. Kozik, D.J. Kliebenstein, R.W. Doerge, D.A. St. Clair, and R.W. Michelmore. 2006. High-density haplotyping with microarray-based expression and single feature polymorphism markers in Arabidopsis. Genome Research. 16:787-795.
D.J. Kliebenstein, M.A.L. West, H. van Leeuwen, O. Loudet, R.W. Doerge, D.A. St Clair. 2006. Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics. 7(1):308.
M.A.L. West, K. Kim, D.J. Kliebenstein, H. van Leeuwen, R.W. Michelmore, R.W. Doerge, D.A. St.Clair. 2007. Global eQTL Mapping Reveals the Complex Genetic Architecture of Transcript Level Variation in Arabidopsis. Genetics. 175: 1441-1450.
H. van Leeuwen, D.J. Kliebenstein, M.A.L. West, K. Kim, R. van Poecke, F. Katagiri, R.W. Michelmore, R.W. Doerge, D.A. St.Clair. 2007. Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. The Plant Cell. 19(7):2099-2110.
Kim, Kyunga. 2007. Statistical Issues in Mapping Genetic Determinants for Expression Level Polymorphisms. Ph.D. Dissertation. Purdue University, West Lafayette, IN 47906 USA