Combining Genomic Data
Meta-analysis is the analysis of analyses, or the combining of results from multiple, similar studies to arrive at a clearer understanding of some underlying effect. When multiple laboratories have conducted experiments using microarrays to study the genetic basis for the same disease or condition of interest, their results will differ due to chance variability as well as fundamental (and sometimes unknown) differences between experiments. For example, each laboratory's list of candidate genes believed to be significantly differentially expressed will differ. A meta-analytic approach to genomic data combines the results from the multiple laboratories in a systematic way to better understand the behavior of individual genes and the sources of differences between the experimental results from the multiple laboratories. Some of the statistical issues involved are accounting for inter-experiment variability, accounting for dependence among experiments from the same lab, and the efficient implementation of a Bayesian approach.
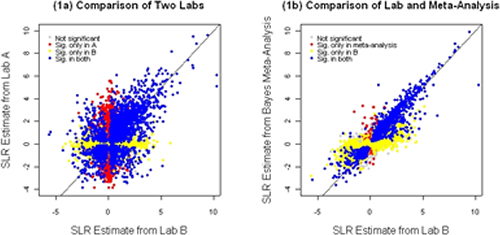
Figure 1. A comparison of the estimates of magnitude of differential expression from different labs and a Bayesian approach to meta-analysis. The SLR (signal log ratio) is the measure of differential expression employed by the Affymetrix MAS 5.0 algorithm. This example represents data collected in multiple labs studying experimental autoimmune encephalomyelitis (EAE) in mouse, with each lab seeking to identify genes differentially expressed between healthy and diseased tissue. The different colors in this figure indicate in which of the labs (and meta-analysis) the individual genes were declared statistically significantly differentially expressed. (a) A comparison of two labs shows considerable variability across labs, due to chance variability and fundamental differences. (b) The meta-analysis takes into account known differences between labs and allows for inter-experiment variability. The meta-analysis then systematically combines results across laboratories to arrive at an overall estimate of the underlying degree of differential expression for each gene as well as a declaration of significance.
References:
Jung Kyoon Choi, Ungsik Yu, Sangsoo Kim, and Ook Joon Yoo. Combining Multiple Microarray Studies and Modeling Interstudy Variation. Bioinformatics, 19(Suppl. 1):i84-i90, 2003.
Giovanni Parmigiani, Elizabeth S. Garrett-Mayer, Ramaswamy Anbazhagan, and Edward Gabrielson. A Cross-Study Comparison of Gene Expression Studies for the Molecular Classification of Lung Cancer. Clinical Cancer Research, 10:2922-2927, 2004.
Ronglai Shen, Debashis Ghosh, and Arul M. Chinnaiyan. Prognostic Meta-Signature of Breast Cancer Developed by Two-Stage Mixture Modeling of Microarray Data. BMC Genomics, 5:94, 2004.
John R. Stevens and R. W. Doerge. Combining Affymetrix Microarray Results. BMC Bioinformatics, accepted March 2005.