Sequencing Analysis
Sequencing of the human genome is mostly complete. Now that we know the basepair composition of the majority of our genome we would like to learn as much as possible from this rich source of data.
These data represent a wealth of information that is difficult to understand and utilize through traditional experimental approaches. To overcome these limitations, statistical methods and probabilistic models are being used to identify functionally relevant sequence motifs (e.g., patterns) within DNA or protein sequence information. Ongoing research at Purdue University, Department of Statistics and colleagues, is focusing on developing statistical approaches to identify conserved motifs in biological sequence data that are critical for the protein-protein or protein-nucleic acid interactions that are central to the fundamental sources of control in biology.
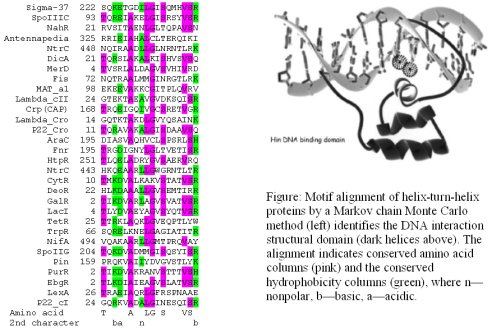
Reference: Xie, J. and Kim, N.K., 2004 (left side graphics), and Feng, J.A., Johnson, R.C. and Dickerson, R.E., 1994 (right side graphics).
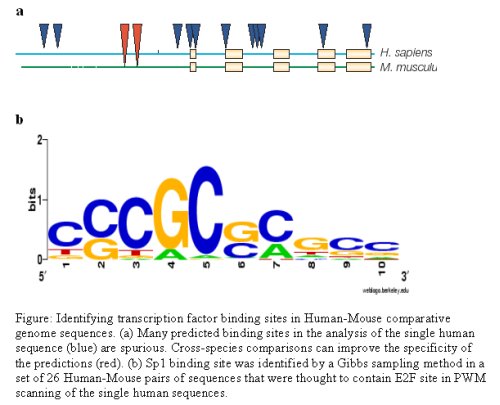
Reference: figure (a) Wasserman, W.W. and Sandelin, A., 2004, figure (b) Xie, J. and Wu, J., 2004