Protein Array Analysis
The past twenty years have witnessed the growth of DNA microarray technology and its important influence in the development of genome discovery. However, the use of transcript profiling is limited in the sense that it only measures transcript abundance at the messenger RNA level.
Studying the genome at the protein level has become the core of many research areas, and is the next step in understanding gene function. There are so many proteins that exist, that when one considers their complex relationships within any one organism, a high-throughput systematic approach for protein analysis is inevitable. Protein microarrays are gaining popularity, and are able to measure the abundance of proteins in a high-throughput manner that is similar to DNA microarrays. Because protein (3-dimensional) structures are significantly different from DNA structures, the design and statistical analysis of protein microarrays is quite different from that of DNA microarrays. To aid in the development of protein microarray technology, our ongoing work includes finding functionally consistent proteins spotted on the microarrays and detecting differentially expressed proteins.
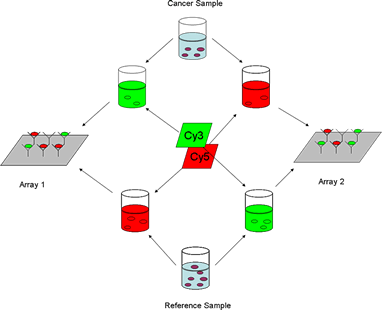
Schematic representation of dye-swap antibody microarray experiment (Miller et al., 2003). The fluorescently labeled proteins of one serum sample of cancer patient were mixed with the fluorescently labeled proteins of a reference sample that is a pool of equal volume of each serum samples. The mixture was then incubated on the antibody microarrays.
Reference: Miller, J.C. and Zhou, H. and Kwekel, J. and Cavallo, R. and Burke, J. and Butler, E.B. and Teh, B.S. and Haab, B.B. Antibody microarray profiling of human prostate cancer sera: Antibody screening and identification of potential biomarkers. Proteomics, 2003, 3:56-63.
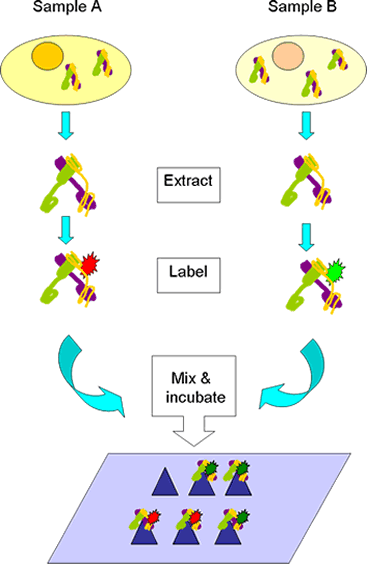
Schematic representation of antibody microarray experiments (Screekumar et al., 2001). Cy5 labeled proteins of LoVo cells was mixed with Cy3 labeled proteins of LoVo cells treated with ionizing radiation, and incubated on the antibody microarray.
Reference: Sreekumar, A. and Nyati, M.K. and Varambally, S. and Barrette, T.R. and Ghosh, D. and Lawrence, S. and Chinnaiyan, A.M. Profilling of cancer cells using protein microarrays: Discovery of novel radiation-regulated proteins. Cancer Research, 2001, 61:7585-7593.
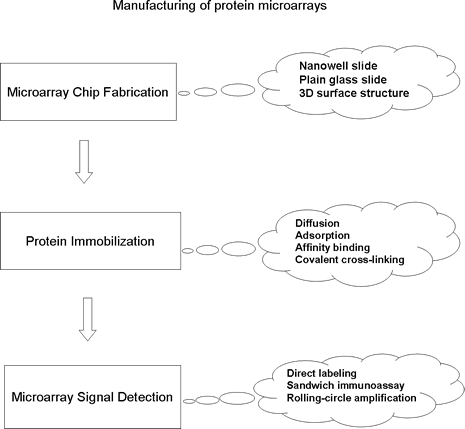
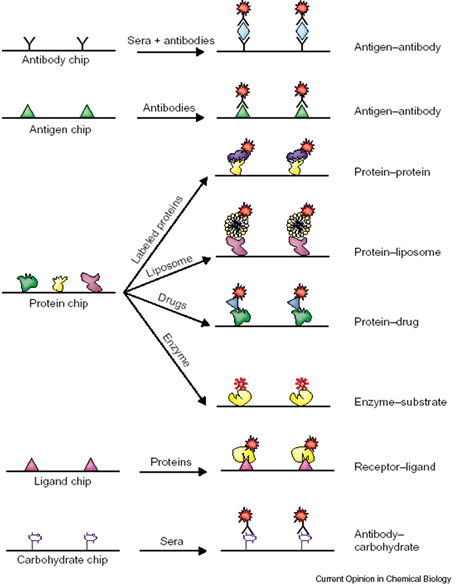
Applications of protein microarrays. There are two general types of protein microarray: analytical and functional protein microarrays. Analytical protein microarrays involve a high-density array of affinity reagents (e.g. antibodies or antigens) that are used for detecting proteins in a complex mixture. Functional protein chips are constructed by immobilizing large numbers of purified proteins on a solid surface. Unlike the antibody antigen chips, protein chips have an enormous potential in assaying for a wide range of biochemical activities (protein-protein, protein-lipid, protein-nucleic acid, and enzyme-substrate interactions), as well as drug and drug target identification. Small molecule and carbohydrate microarrays are other types of analytical microarrays that have been demonstrated to be capable of studying protein binding activities to ligands and carbohydrates.
Reference: Zhu, H. and Snyder, M. Protein chip technology. Current Opinion in Chemical Biology, 2003, 7:55-63.